EOSC-Life

The EOSC-Life project develops an open collaborative space for digital biology in Europe
- Website: http://www.eosc-life.eu/
- Started: March 1, 2019
- Duration: 54 months
- Project reference: https://doi.org/10.3030/824087
EOSC-Life brought together the 13 Biological and Medical ESFRI research infrastructures (BMS RIs) to create an open collaborative space for digital biology. This was a joint response to the challenge of analysing and reusing the prodigious amounts of data produced by life-science. Managing and integrating this data is beyond the capabilities of most individual end-users and institutes. By publishing data and tools in a Europe-wide cloud, EOSC-Life aimed to bring the capabilities of big science projects to the wider research community. Federated user access (AAI) allows transnational resource access and authorisation. EOSC-Life establishes a novel access model for the BMS RI: through EOSC scientists would gain direct access to FAIR data and tools in a cloud environment available throughout the European Research Area.
Funded by call H2020-INFRAEOSC-2018-2 824087, EOSC-Life made BMS RIs data resources FAIR and publish them in the EOSC following guidelines and standards (e.g. EDMI). Overall this drive the evolution of the RI repository infrastructure for EOSC and integration of the BMS RI repositories. EOSC-Life implemented workflows that cross disciplines and RI boundaries and addressed the needs of interdisciplinary science. Through open hackathons and bring-your-own-data events EOSC-Life was co-created with exustubg user communities, providing a blueprint for how the EOSC could support wide-spread and excellent data-driven life science research. EOSC-Life started adressing the data policies needed for human research data under GDPR. Interoperable provenance information describe history of sample and data to ensure reproducibility and adherence to regulatory requirements.
The goal of the EOSC-Life project was to make sure that life-scientists can find, access and integrate life-science data for analysis and reuse in academic and industrial research. EOSC-Life will transform European life-science by providing an open, continent-scale, collaborative and interdisciplinary environment for data science.
Subsequent projects building on EOSC-Life technologies and communities includes:
- BY-COVID – building on EOSC-Life’s provenance work with Workflow Run Crate and Common Provenance Model.
- EOSC4Cancer – reusing workflow approach for federated EOSC services for cancer research
- EuroScienceGateway – extending Galaxy’s RO-Crate support and linking to FAIR Digital Objects
- TRE-FX – using WfExS workflow execution service and RO-Crate for federated workflow execution across trusted research environments
eScienceLab contribution
The eScience Lab contributions in eScienceLab were part of ELIXIR-UK.
- WP1: Publishing FAIR RI data resources in EOSC
- WP2: Tools Collaboratory: Deployment of life-science data integration and analysis workflows in EOSC
- Implementation of a mechanism for publishing and sharing workflows across instances of the environment
- WP3: Demonstrators and Open Calls for User Projects
We addressed cross-RI interoperability in the EOSC by fostering implementation of tools and workflows in an integrated EOSC environment. We leveragde pre-existing work on standardization synergized with relevant efforts such as GA4GH Cloud Work Stream.
The integrated environment is built on:
- software and tools packaging will leverage existing efforts around package managers for software (e.g. BioConda) and containerization technology (e.g. Docker, Singularity) with reference to standards from the Open Containers Initiative.
- workflow composition and execution will rely on widely adopted standards for workflow specification (e.g. Common Workflow Language) to ensure interoperability and re-usability across research infrastructure.
- an environment with a graphical user interface (e.g. based on Galaxy) to support end-users with little or no experience in the design, customization and implementation of software pipelines.
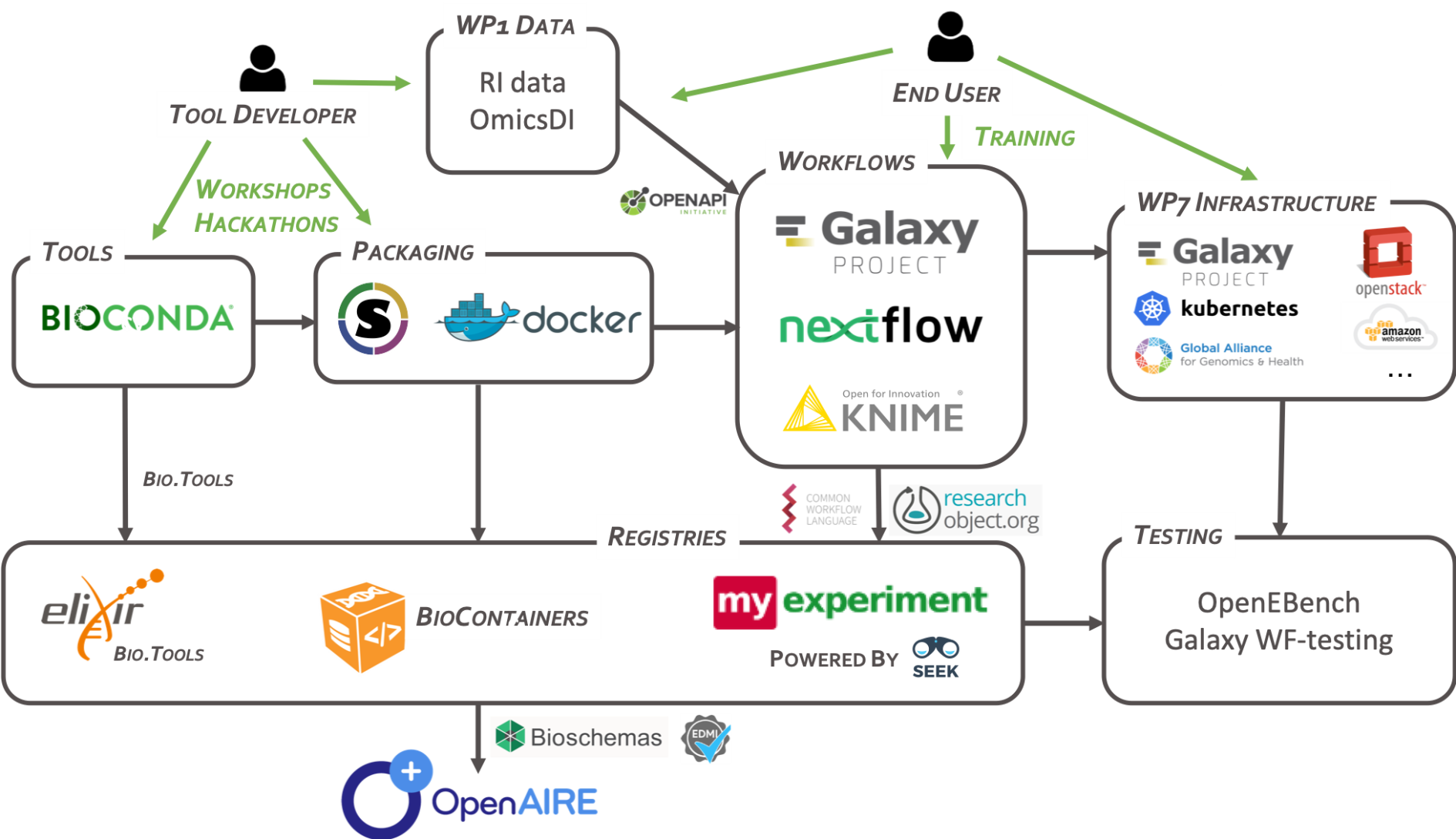
Tools and workflows must be findable and accessible. They should also be citable, have managed metadata profiles and openly available for review and analytics. The registry solution has these components:
- WorkflowHub, a Workflow Registry that is workflow system agnostic, supports a repository of workflows in native and standardised form (e.g. CWL) and the virtual aggregation of established tool, workflow and registries to support discovery over a fragmented ecosystem. The federated registry support a common API to simplify access for tool developers.
- Standardised workflow identifiers and metadata descriptions (needed for workflow discovery, reuse, preservation, interoperability and monitoring and metadata harvesting using standard protocols. Workflows are usually multi-component (requiring links to test data, example runs, explanatory documentation, etc) and used in collections for scientific use cases.
- We matured RO-Crate and developed several profiles for workflows: Workflow RO-Crate (which is the encapsulation used by WorkflowHub API) and Workflow Run RO-Crate (which captures the provenance recording of a particular workflow run, implemented by multiple workflow engines)
- Workflow snapshot preservation, publishing, citation and monitoring, credit claiming and workflows part of the scholarly communication landscape partnering DataCite and EOSC’s OpenAIRE, linking publications with workflows, associated datasets, software. WorkflowHub uses DOIs for citable workflows.
WorkflowHub is based on the SEEK platform using Common Workflow Language and Research Objects to glue in federated workflow and tool descriptions across the research infrastructures. It is integrated with the Life Science Login, GA4GH TRS API, European Galaxy service, LifeMonitor, ELIXIR’s Tool registry bio.tools and other services. WorkflowHub is an EOSC service.
Related pages
- EOSC-Life homepage
- EOSC-Life Tools & Workflows
- ELIXIR: The EOSC-Life project develops an open collaborative space for digital biology in Europe
Related publications
For a complete list, see EOSC-Life Publications.
Romain David, Arina Rybina, Jean‐Marie Burel, Jean‐Karim Heriche, Pauline Audergon, Jan‐Willem Boiten, Frederik Coppens, Sara Crockett, Katrina Exter, Sven Fahrner, Maddalena Fratelli, Carole Goble, Philipp Gormanns, Tobias Grantner, Björn Grüning, Kim Tamara Gurwitz, John M Hancock, Henriette Harmse, Petr Holub, Nick Juty, Geoffrey Karnbach, Emma Karoune, Antje Keppler, Jessica Klemeier, Carla Lancelotti, Jean‐Luc Legras, Allyson L Lister, Dario Livio Longo, Rebecca Ludwig, Bénédicte Madon, Marzia Massimi, Vera Matser, Rafaele Matteoni, Michaela Th Mayrhofer, Christian Ohmann, Maria Panagiotopoulou, Helen Parkinson, Isabelle Perseil, Claudia Pfander, Roland Pieruschka, Michael Raess, Andreas Rauber, Audrey S Richard, Paolo Romano, Antonio Rosato, Alex Sánchez‐Pla, Susanna‐Assunta Sansone, Ugis Sarkans, Beatriz Serrano‐Solano, Jing Tang, Ziaurrehman Tanoli, Jonathan Tedds, Harald Wagener, Martin Weise, Hans V Westerhoff, Rudolf Wittner, Jonathan Ewbank, Niklas Blomberg, Philip Gribbon (2023):
“Be sustainable”: EOSC‐Life recommendatons for implementation of FAIR principles in life science data handling.
The EMBO Journal 42:e115008
https://doi.org/10.15252/embj.2023115008
Rudolf Wittner, Petr Holub, Cecilia Mascia, Francesca Frexia,
Heimo Müller, Markus Plass, Clare Allocca, Fay Betsou,
Tony Burdett, Ibon Cancio, Adriane Chapman, Martin Chapman,
Mélanie Courtot, Vasa Curcin, Johann Eder, Mark Elliot,
Katrina Exter, Carole Goble, Martin Golebiewski,
Bron Kisler, Andreas Kremer, Simone Leo, Sheng Lin-Gibson,
Anna Marsano, Marco Mattavelli, Josh Moore, Hiroki Nakae,
Isabelle Perseil, Ayat Salman, James Sluka,
Stian Soiland-Reyes, Caterina Strambio-De-Castillia,
Michael Sussman, Jason R. Swedlow, Kurt Zatloukal, Jörg Geiger (2023):
Towards a common standard for data and specimen provenance in life sciences.
Learning Health Systems
https://doi.org/10.1002/lrh2.10365
Romain David, Audrey S. Richard, Claire Connellan, Katharina B. Lauer, Maria Luisa Chiusano, Carole Goble, Martin Houde, Isabel Kemmer, Antje Keppler, Philippe Lieutaud, Christian Ohmann, Maria Panagiotopoulou, Sara Raza Khan, Arina Rybina, Stian Soiland-Reyes, Charlotte Wit, Rudolf Wittner, Rafael Andrade Buono, Sarah Arnaud Marsh, Pauline Audergon, Dylan Bonfils, Jose-Maria Carazo, Remi Charrel, Frederik Coppens, Wolfgang Fecke, Claudia Filippone, Eva Garcia Alvarez, Sheraz Gul, Henning Hermjakob, Katja Herzog, Petr Holub, Lukasz Kozera, Allyson L. Lister, José López-Coronado, Bénédicte Madon, Kurt Majcen, William Martin, Wolfgang Müller, Elli Papadopoulou, Christine M.A. Prat, Paolo Romano, Susanna-Assunta Sansone, Gary Saunders, Niklas Blomberg, Jonathan Ewbank (2023):
Umbrella Data Management Plans to integrate FAIR data: lessons from the ISIDORe and BY-COVID consortia for pandemic preparedness.
Zenodo
https://doi.org/10.5281/zenodo.7520086
Stian Soiland-Reyes, Leyla Jael Castro, Daniel Garijo, Marc Portier, Carole Goble, Paul Groth (2022):
Updating Linked Data practices for FAIR Digital Object principles.
Research Ideas and Outcomes 8:e94501
https://doi.org/10.3897/rio.8.e94501
Paul De Geest, Frederik Coppens, Stian Soiland-Reyes, Ignacio Eguinoa, Simone Leo (2022):
Enhancing RDM in Galaxy by integrating RO-Crate.
Research Ideas and Outcomes 8:e95164
https://doi.org/10.3897/rio.8.e95164
Stian Soiland-Reyes, Peter Sefton, Leyla Jael Castro, Frederik Coppens, Daniel Garijo, Simone Leo, Marc Portier, Paul Groth (2022):
Creating lightweight FAIR Digital Objects with RO-Crate.
Research Ideas and Outcomes 8:e93937
https://doi.org/10.3897/rio.8.e93937
Fuqi Xu, Nick Juty, Carole Goble, Simon Jupp, Helen Parkinson, Mélanie Courtot (2022):
Features of a FAIR vocabulary.
Journal of Biomedical Semantics 14:6
https://doi.org/10.1186/s13326-023-00286-8
Michael R. Crusoe, Sanne Abeln, Alexandru Iosup, Peter Amstutz, John Chilton, Nebojša Tijanić, Hervé Ménager, Stian Soiland-Reyes, Bogdan Gavrilović, Carole Goble, The CWL Community (2022):
Stian Soiland-Reyes, Peter Sefton, Mercè Crosas, Leyla Jael Castro, Frederik Coppens, José M. Fernández, Daniel Garijo, Björn Grüning, Marco La Rosa, Simone Leo, Eoghan Ó Carragáin, Marc Portier, Ana Trisovic, RO-Crate Community, Paul Groth, Carole Goble (2022):
Packaging research artefacts with RO-Crate.
Data Science 5(2)
https://doi.org/10.3233/DS-210053
Rudolf Wittner, Cecilia Mascia, Matej Gallo, Francesca Frexia, Heimo Müller, Markus Plass, Jörg Geiger & Petr Holub (2022):
Lightweight Distributed Provenance Model for Complex Real–world Environments.
Scientific Data 9:503
https://doi.org/10.1038/s41597-022-01537-6
Francesca Frexia, Cecilia Mascia, Rudolf Wittner, Markus Plass, Heimo Müller, Jörg Geiger, Petr Holub (2022):
The Common Provenance Model: Capturing Distributed Provenance in Life Sciences Processes.
In: Brigitte Séroussi, Patrick Weber, Ferdinand Dhombres, Cyril Grouin, Jan-David Liebe, Sylvia Pelayo, Andrea Pinna, Bastien Rance, Lucia Sacchi, Adrien Ugon, Arriel Benis, Parisis Gallos (eds.) Challenges of Trustable AI and Added-Value on Health
Studies in Health Technology and Informatics 294
https://doi.org/10.3233/SHTI220489
Michael R. Crusoe, Sanne Abeln, Alexandru Iosup, Peter Amstutz, John Chilton, Nebojša Tijanić, Hervé Ménager, Stian Soiland-Reyes, Bogdan Gavrilović, Carole Goble, The CWL Community (2022):
Methods Included: Standardizing Computational Reuse and Portability with the Common Workflow Language.
Communications of the ACM 65(6)
https://doi.org/10.1145/3486897
Paul Brack, Peter Crowther, Stian Soiland-Reyes, Stuart Owen, Douglas Lowe, Alan R Williams, Quentin Groom, Mathias Dillen, Frederik Coppens, Björn Grüning, Ignacio Eguinoa, Philip Ewels, Carole Goble (2022):
Ten Simple Rules for making a software tool workflow-ready.
PLOS Computational Biology 18(3):e1009823
https://doi.org/10.1371/journal.pcbi.1009823
Stian Soiland-Reyes, Genís Bayarri, Pau Andrio, Robin Long, Douglas Lowe, Ania Niewielska, Adam Hospital, Paul Groth (2022):
Making Canonical Workflow Building Blocks interoperable across workflow languages.
Data Intelligence 4(2)
https://doi.org/10.1162/dint_a_00135
Peter Wittenburg, Ivonne Anders, Christophe Blanchi, Merret Buurman, Carole Goble, Jonas Grieb, Alex Hardisty, Sharif Islam, Thomas Jejkal, Tibor Kálmán, Christine Kirkpatrick, Laurence Lannom, Thomas Lauer, Giridhar Manepalli, Karsten Peters-von Gehlen, Andreas Pfeil, Robert Quick, Mark van de Sanden, Ulrich Schwardmann, Stian Soiland-Reyes, Rainer Stotzka, Zachary Trautt, Dieter Van Uytvanck, Claus Weiland, Philipp Wieder (2022):
FAIR Digital Object Demonstrators 2021.
Report, FAIR Digital Objects Forum. Zenodo
https://doi.org/10.5281/zenodo.5872645
Anna-Lena Lamprecht, Magnus Palmblad, Jon Ison, Veit Schwämmle,
Mohammad Sadnan Al Manir, Ilkay Altintas, Christopher J. O. Baker, Ammar Ben Hadj Amor, Salvador Capella-Gutierrez,
Paulos Charonyktakis, Michael R. Crusoe,
Yolanda Gil, Carole Goble, Timothy J. Griffin,
Paul Groth, Hans Ienasescu, Pratik Jagtap,
Matúš Kalaš, Vedran Kasalica, Alireza Khanteymoori,
Tobias Kuhn, Hailiang Mei, Hervé Ménager, Steffen Möller, Robin A. Richardson,
Vincent Robert, Stian Soiland-Reyes, Robert Stevens, Szoke Szaniszlo,
Suzan Verberne, Aswin Verhoeven, Katherine Wolstencroft (2021):
Perspectives on automated composition of workflows in the life sciences.
[version 1; peer review: 2 approved]
F1000Research 10:897
https://doi.org/10.12688/f1000research.54159.1
Rafael Ferreira da Silva, Henri Casanova, Kyle Chard, Ilkay Altintas, Rosa M Badia, Bartosz Balis, Tainã Coleman, Frederik Coppens, Frank Di Natale, Bjoern Enders, Thomas Fahringer, Rosa Filgueira, Grigori Fursin, Daniel Garijo, Carole Goble, Dorran Howell, Shantenu Jha, Daniel S. Katz, Daniel Laney, Ulf Leser, Maciej Malawski, Kshitij Mehta, Loïc Pottier, Jonathan Ozik, J. Luc Peterson, Lavanya Ramakrishnan, Stian Soiland-Reyes, Douglas Thain, Matthew Wolf (2021):
A Community Roadmap for Scientific Workflows Research and Development.
2021 IEEE Workshop on Workflows in Support of Large-Scale Science (WORKS), pp 81–90.
https://doi.org/10.1109/WORKS54523.2021.00016
arXiv:2110.02168 [cs.DC]
Carole Goble, Stian Soiland-Reyes, Finn Bacall, Stuart Owen, Alan Williams, Ignacio Eguinoa, Bert Droesbeke, Simone Leo, Luca Pireddu, Laura Rodriguez-Navas, José Mª Fernández, Salvador Capella-Gutierrez, Hervé Ménager, Björn Grüning, Beatriz Serrano-Solano, Philip Ewels, Frederik Coppens (2021):
Implementing FAIR Digital Objects in the EOSC-Life Workflow Collaboratory.
Zenodo
https://doi.org/10.5281/zenodo.4605654
Rudolf Wittner, Petr Holub, Heimo Müller, Jörg Geiger, Carole Goble, Stian Soiland-Reyes, Luca Pirredu, Francesca Frexia, Cecilia Mascia, Elliot Fairweather, Jason R. Swedlow, Josh Moore, Caterina Strambio, Gianluigi Zanetti, David Grunwald, Hiroki Nakae (2021):
ISO 23494: Biotechnology - Provenance Information Model for Biological Specimen and Data.
In: Provenance and Annotation of Data and Processes. IPAW 2020, IPAW 2021.
Lecture Notes in Computer Science 12839
https://doi.org/10.1007/978-3-030-80960-7_16 [preprint]
Carole Goble, Sarah Cohen-Boulakia, Stian Soiland-Reyes, Daniel Garijo, Yolanda Gil, Michael R. Crusoe, Kristian Peters, Daniel Schober (2020):
FAIR Computational Workflows.
Data Intelligence 2(1):108–121
https://doi.org/10.1162/dint_a_00033
Elliot Fairweather, Rudolf Wittner, Martin Chapman, Petr Holub, Vasa Curcin (2020):
Non-repudiable provenance for clinical decision support systems.
arXiv:2006.11233 [cs.CR]
https://doi.org/10.48550/arXiv.2006.11233
Annika Jacobsen, Ricardo de Miranda Azevedo, Nick Juty, Dominique Batista, Simon Coles, Ronald Cornet, Mélanie Courtot, Mercè Crosas, Michel Dumontier, Chris T. Evelo, Carole Goble, Giancarlo Guizzardi, Karsten Kryger Hansen, Ali Hasnain, Kristina Hettne, Jaap Heringa, Rob W.W. Hooft, Melanie Imming, Keith G. Jeffery, Rajaram Kaliyaperumal, Martijn G. Kersloot, Christine R. Kirkpatrick, Tobias Kuhn, Ignasi Labastida, Barbara Magagna, Peter McQuilton, Natalie Meyers, Annalisa Montesanti, Mirjam van Reisen, Philippe Rocca-Serra, Robert Pergl, Susanna-Assunta Sansone, Luiz Olavo Bonino da Silva Santos, Juliane Schneider, George Strawn, Mark Thompson, Andra Waagmeester, Tobias Weigel, Mark D. Wilkinson, Egon Willighagen, Peter Wittenburg, Marco Roos, Barend Mons, Erik Schultes (2020):
FAIR Principles: Interpretations and Implementation Considerations.
Data Intelligence 2(1):10–29
https://doi.org/10.1162/dint_r_00024
Mark D. Wilkinson, Michel Dumontier, Susanna-Assunta Sansone, Luiz Olavo Bonino da Silva Santos, Mario Prieto, Dominique Batista, Peter McQuilton, Tobias Kuhn, Philippe Rocca-Serra, Mercѐ Crosas, Erik Schultes (2019):
Evaluating FAIR maturity through a scalable, automated, community-governed framework.
Scientific Data 6(1)
https://doi.org/10.1038/s41597-019-0184-5
Related deliverables
For a complete list, see EOSC-Life Project deliverables.
Niklas Blomberg, Niklas, Friederike Schmidt-Tremmel, Sara Crockett (2023):
EOSC-Life Summary of EOSC-Life Main Achievements and Impacts with Sustainability and Exploitation Plans.
EOSC-Life, Project deliverable D11.3
https://doi.org/10.5281/zenodo.8304575
Maddalena Fratelli, Gary Saunders, Arina Rybina, Jing Tang (2023):
EOSC-Life Report on reproducibility.
EOSC-Life, Project deliverable D8.4
https://doi.org/10.5281/zenodo.8300699
Rudolf Wittner, Jörg Geiger, Heimo Müller, Francesca Frexia, Cecilia Mascia, Katrina Exter, Ibon Cancio, Petr Holub (2023):
EOSC-Life Common provenance model for processing biological material, data generation and computational workflows.
EOSC-Life, Project deliverable D6.6
https://doi.org/10.5281/zenodo.8279525
Arina Rybina, Pauline Audergon, Claudia Pfander (2023):
EOSC-Life Report on the work of the Open Call Projects.
EOSC-Life, Project deliverable D3.3
https://doi.org/10.5281/zenodo.8263074
Carole Goble, Stian Soiland-Reyes, Finn Bacall, Stuart Owen, Luca Pireddu, Simone Leo (2023):
EOSC-Life Implementation of a mechanism for publishing and sharing workflows across instances of the environment.
EOSC-Life, Project deliverable D2.3
https://doi.org/10.5281/zenodo.7886545
Antonio Rosato, Jean-Karim Hériché (2022):
EOSC-Life A common environment that can run cross-RIs workflows.
EOSC-Life, Project deliverable D2.2
https://doi.org/10.5281/zenodo.7217294
Florence Bietrix, José Maria Carazo, Salvador Capella-Gutierrez, Frederik Coppens, Maria Luisa Chiusano, Romain David, Jose Maria Fernandez, Maddalena Fratelli, Jean-Karim Heriche, Carole Goble, Philip Gribbon, Petr Holub, Robbie P. Joosten, Simone Leo, Stuart Owen, Helen Parkinson, Roland Pieruschka, Luca Pireddu, Luca Porcu, Michael Raess, Laura Rodriguez-Navas, Andreas Scherer, Stian Soiland-Reyes, Jing Tang, Gary Saunders (2022):
EOSC-Life Methodology framework to enhance reproducibility within EOSC-Life – revised version
EOSC-Life, Project deliverable D8.1 (revised)
https://doi.org/10.5281/zenodo.7137744
Helen Parkinson, Philip Gribbon, Ugis Sarkans, Gesa Witt, Andrea Zaliani, Manfred Kohler, Jason Swedlow, Jean-Marie Burel, Morris Swertz, Esther van Enckevort, Petr Holub, Marzia Massimi, Rafaele Matteoni, Holger Maier, Reetta Hinttala, Anne Heikkinen, Philipp Gormanns, Laurent Vasseur, Sophie Leblanc, Yann Herault, Dimitris Kontoyiannis, Christina Chandras, Dimitra Panou, José Miguel López Coronado, Rosa Aznar Novella, Vincent Robert, Ammar Ben Hadj Amor, Serge Casaregola, Jean-Luc Legras, Michel-Yves Mistou, Paolo Romano, Isabelle Perseil, Romain David, Roland Pieruschka, Katrina Exter, Marc Portier, Cedric Decruw, Steve Canham, Christian Ohmann, Sergey Goryanin, Laura Del Cano, Maddalena Fratelli, Carole Goble, Stuart Owen, Stian Soiland- Reyes, Nick Juty, Henriette Harmse, Dario Longo, Susanna Sansone, Allyson L. Lister, Peter McQuilton, Milo Tursthon, Ramon Granell, Hossein Mirian, Marco Roos, Luiz Bonino (2021):
EOSC-Life EOSC FAIR services deployment for open calls.
EOSC-Life, Project deliverable D1.3
https://doi.org/10.5281/zenodo.6208249
Frauke Leitner, Jose Maria Carazo, Johanna Bischof, Natalie Haley, Pauline Audergon, Carlos Oscar Sorzano, Laura del Cano, Pablo Conesa, Cymon J. Cox, Gianluca De Moro, Corre Erwan, Katrina Exter, Gildas Le Corguillé, Romain Dallet, Lorraine Gueguen, Jean-Karim Heriche, Beatriz Serrano, Yi Sun, Jean-Marie Burel, Sara Zullino, Dario Livio Longo, Cyril Pommier, Asis Hallab, Constantin Eiteneuer, Romain David, Bjorn Usadel, Stuart Owen, Kristina Gruden, Roland Pieruschka, Alexander Sczyrba, Alfred Puhler, Martin Beracochea, Robert Finn, Philip Gribbon, Andrea Zaliani, Rachael Skyner, Frank von Delft, Ctibor Skuta, Andrew Leach, Jing Tang, Salvador Capella, José M. Fernández, Jordi Rambla, Sergi Beltran (2021):
EOSC-Life Report on the work of the initial demonstrators.
EOSC-Life, Project deliverable D3.2
https://doi.org/10.5281/zenodo.4817723
Rudolf Wittner, Cecilia Mascia, Francesca Frexia, Heimo Müller, Jörg Geiger, Katrina Exter & Petr Holub (2021):
EOSC-Life Common Provenance Model.
EOSC-Life, Project deliverable D6.2
https://doi.org/10.5281/zenodo.4705074
Helen Parkinson, Philip Gribbon, Ugis Sarkans, Gesa Witt, Andrea Zaliani, Manfred Kohler, Jason Swedlow, Jean-Marie Burel, Morris Swertz, Esther van Enckevort, Petr Holub, Marzia Massimi, Rafaele Matteonie, Holger Maier, Reetta Hinttala, Anne Heikkinen, Philipp Gormanns, Laura del Cano, Laurent Vasseur, Sophie Leblanc, Yann Herault, Dimitris Kontoyiannis, Christina Chandras, Dimitra Panou, José Miguel López Coronado, Rosa Aznar Novella, Vincent Robert, Ammar Ben Hadj Amor, Serge Casaregola, Jean-Luc Legras, Michel-Yves Mistou, Paolo Romano, Isabelle Perseil, Romain David, Roland Pieruschka, Katrina Exter, Marc Portier, Cedric Decruw, Steve Canham, Christian Ohmann, Sergey Goryanin, Laura Del Cano, Maddalena Fratelli, Carole Goble, Stuart Owen, Stian Soiland-Reyes, Nick Juty, Susanna Sansone, Peter McQuilton, Marco Roos, Luiz Bonino (2021):
EOSC-Life EOSC repository deployment for project demonstrators.
EOSC-Life, Project deliverable D1.2
https://doi.org/10.5281/zenodo.4432029
Antonio Rosato, Jean-Karim Hériché, Beatriz Serrano-Solano, Björn Grüning, Luca Pireddu & Carole Goble (2020):
EOSC-Life Cloud implementation of exemplary workflows.
EOSC-Life, Project deliverable D2.1
https://doi.org/10.5281/zenodo.4010401
